HOME
Slow-growing rotavirus mutant reveals early steps of viral assembly
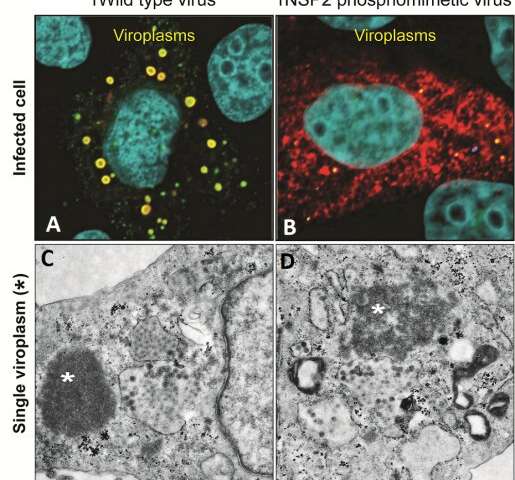
Rotaviruses infect cells to produce more viruses. One of the first steps of rotavirus replication is building rotavirus factories called viroplasms, seen in panel A as round, yellow dots inside rotavirus-infected monkey kidney epithelial cells (cell nuclei are colored in teal). But when rotaviruses carry the NSP2 mutant protein (shown in red), they are delayed in forming viroplasms, as shown by the presence of few, smaller yellow dots in panel B. The bottom panels are electron micrographs of a single viroplasm (marked by *). Panel C shows that typically viroplasms are dense rounded structures. In contrast, as shown in panel D, rotaviruses carrying mutant protein NSP2 form viroplasms with a disorganized architecture. Credit: J. Criglar/Estes lab/J. Virology, 2020
Rotavirus is responsible for more than 130,000 deaths in infants and young children younger than five years, every year. The virus causes severe, dehydrating diarrhea as it replicates in viral factories called viroplasms that form inside infected cells. Viroplasms have been difficult to study because they normally form very quickly, but a serendipitous observation led researchers at Baylor College of Medicine to uncover new insights into the formation of viroplasms.
The researchers created a mutant rotavirus that unexpectedly replicated much slower than the original virus, allowing them to observe the first steps of viral assembly. The findings, published in the Journal of Virology, open new possibilities for treating and preventing this viral disease and for understanding how similar factories of other viruses work. "The formation of viroplasms is indispensable for a successful rotavirus infection. They form quickly inside infected cells and are made of both viral and cellular proteins that interact with lipid droplets, but the details of how the parts are put together are still not clear," said first author Dr. Jeanette M. Criglar, a former postdoctoral trainee and now staff scientist in the Department of Molecular Virology and Microbiology at Baylor in Dr. Mary Estes's lab.
To get new insights into the formation of viroplasms, Criglar and her colleagues studied NSP2, one of the viral proteins that is required for the virus to replicate. Without it, neither viroplasms nor new viruses would form. Like all proteins, NSP2 is made of amino acids strung together like beads on a necklace. 'Bead' 313 is the amino acid serine. Importantly, serine 313 is phosphorylated—it has a phosphate chemical group attached to it. Protein phosphorylation is a mechanism cells use to regulate protein activity. It works like an on-and-off switch, activating or deactivating a protein. Here, the researchers evaluated the role NSP2's phosphorylation of serine 313 plays on viroplasm formation.
News Source